By Ethan Covey
Researchers have used genomic sequencing to highlight the connection among identified cases of blastomycosis, according to a recent study (Emerg Infect Dis 2022;28[9]:1924-1926).
“This study is the first to use genomic sequencing for an outbreak investigation of blastomycosis in the U.S.,” said Nancy A. Chow, PhD, the data and quality team leader for the Mycotic Diseases branch in the Division of Foodborne, Waterborne and Environmental Diseases, at the CDC, in Atlanta.
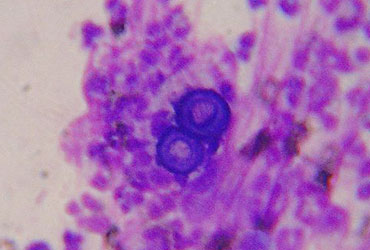
The report focused on two outbreaks of blastomycosis, which occurred in Minnesota.
In the first outbreak (cluster A), five cases from a single family were identified, two white-Hispanic parents and three children. In the second cluster (cluster B), two cases of blastomycosis were identified in white non-Hispanic sisters.
Whole-genome sequencing was performed to determine the genetic diversity and phylogenetic relationships of the two clusters.
All isolates were found to be Blastomyces gilchristii rather than Blastomyces dermatitidis, and the cases within cluster A or B were determined to be closely related genetically, whereas clusters A and B were genetically distinct.
“The genomic findings corresponded with the epidemiological information where the strains from cases within a family cluster were more closely related than the strains from cases between family clusters,” Dr. Chow said. “This work highlights the utility of an advanced molecular technology to conduct genomic epidemiology and surveillance of this disease.”
Dr. Chow added that public health would benefit from future research focused on describing the geographic distribution of Blastomyces species in the United States, and how the genetic diversity of species differs by geography.
“Additionally, it is critical to understand whether, and if so, how climate change may affect the geographic distribution of Blastomyces species in the U.S.,” she said.
